Create new variables
Now we can extend the dataset QldShark_2017_Clean_v2 with a new variable Rainfall_mm from the BOM_GCS_Rain_2017 dataset. We will do this using a GREL expression cell.cross.
The cell.cross function performs a cross or lookup between two columns in two datasets or the same dataset. It returns an array (list) of zero or more rows in the project i.e. matching against BOM_GCS_Rain_2017 for which the cells in the column i.e. Date have the same content as cell Date .
Match a key variable and add a new variable using GREL cell.cross
- Go to
QldShark_2017_Clean_v2project Firstly let’s transform theDatecolumn to a date format. - Select
Datecolumn > Edit cells > Common transforms > To date`
Now
- Select
Areacolumn> Facet > Text Facet - Select
Gold Coastfrom Text Facet results menu
56 resulting matching rows will display which you can now work with.
- On
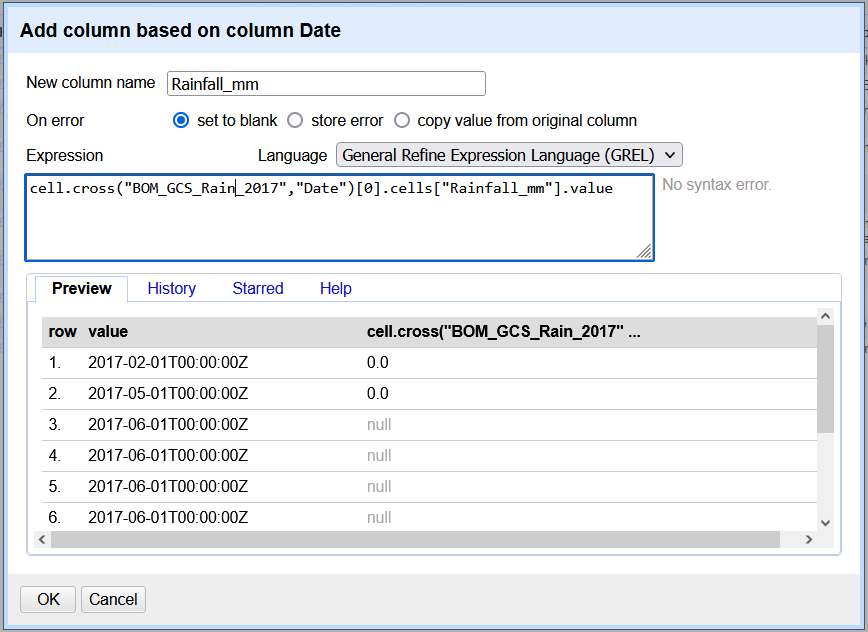
DateselectEdit Column > Add column based on this column - name the new Column
Rainfall_mm -
enter this GREL expression:
cell.cross("BOM_GCS_Rain_2017","Date")[0].cells["Rainfall_mm"].value("BOM_GCS_Rain_2017","Date")is the data we are looking up and matching.[0]counts from the first value..cells[“Rainfall_mm”].valueis a command to add the value from the Rainfall_mm variable to the new column, if there is a match.
- 56 rows of the shark captures now have a new variable & value of rainfall added.
xclose the Text Facet window.
The image below shows the GREL cell.cross expression in action.
Follow the steps in this video.
Let’s now extend the dataset QldShark_2017_Clean_v2 with another variable Species from the NESP_SharkSpeciesList.csv dataset.
Tidy the data then match a key variable using GREL cell.cross
QldShark_2017_Clean_v2 currently contains a variable Species Name , but the values are actually common names the sharks are known by. Let’s change the variable name.
- Go to
QldShark_2017_Clean_v2project - Select
Species Namecolumn> Edit column > Rename this column - name it
Common name Ok.
Next:
- Select the Open button in the top right hand corner of OpenRefine to open a new instance of the software in your browser.
- Create a new project using
NESP_SharkSpeciesList.csvfile - Name it
NESP_SharkSpeciesList - Look at both datasets, what variables do they have in common?
Common namein ``NESP_SharkSpeciesList.csv`Common nameinNESP_SharkSpeciesList.
- Go to
Common namefor each dataset Facet > Text facet
Take a look at the names in each. The NESP_SharkSpeciesList dataset contains an authoritative list of Shark common names, Species, Genus, Family and Order names from a scientific project. The QldShark_2017_Clean_v2 list of names is not authoritative, and many of the shark common names are variations or incorrect. We need to correct the following:
All the “Whaler” names in QldShark_2017_Clean_v2 need to be changed to “Shark” as the only Whalers identified in NESP_SharkSpeciesList were Bronze and Creek Whaler. You can perform a filter on whaler to check.
We need to clean the inconsistent names to then match the values in the key variable accurately.
- Go to
Common nameinQldShark_2017_Clean_v2 Edit cells > Replace- Find:
whaler - tick
case insensitive - Replace with:
Shark Ok.
Then on the same column:
Facet > Text Facet
The first shark name is an abbreviation let’s make it a full name.
- highlight
Aus Sharpnose Shark>Edit - Change to
Australian Sharpnose Shark>Apply - Leave the
Text facetwindow open.
We want to match on unique values in the variables, and at present many of the shark names in QldShark_2017_Clean_v2 are missing the term “Shark” in their title. We can check them against the Common name’s in NESP_SharkSpeciesList.
- Look through the
Text Facetlist inQldShark_2017_Clean_v2for names withoutSharkin their name. These include: -
Australian Blacktip,Great Hammerhead,MakoandWobbegong - Go to
Common nameinNESP_SharkSpeciesList>Text filter - type
Australian Blacktip- note the results that the common name needs
Sharkadded. Make this change inQldShark_2017_Clean_v2
- note the results that the common name needs
- now type
Great Hammerheadin theText filter- note the results are the same, so no changes needed
- perform a filter for each of the four names above and check if correctly named.
- note that
MakoandWobegongare generic names and we can’t identify the species exactly, so we won’t be able to match on these.
Now that we have cleaned up as many inconsistancies possible, let’s match the key variables:
- Go to
QldShark_2017_Clean_v2project - at
Common nameselectEdit Column > Add column based on this column - name the new Column
Species -
enter this GREL expression: (tip: you can reuse and make changes to previous expressions using the
Historytab)cell.cross("NESP_SharkSpeciesList","Common name")[0].cells["Species"].value - Preview and
ok.
382 rows of the shark captures now have a new variable & value of Species added.
xclose the Text Facet window.
Challenge: Try adding the Genus variable using the same steps as above.
Find more information on the cell.cross function here and more GREL functions here.